Random walk vectors for clustering (part I – similarity between objects)
This post opens a series of short discussions on how to use multiple random walk vectors (vectors describing probability distributions of a random surfer on a graph—like PageRank) to find homogeneous groups of objects in a dataset. The main idea here is to combine the existing work of many researchers into one method. The mathematics behind these ideas might be complex, but we will try to be as ELI5-oriented as possible—so anyone can read, understand (at least the basics), and implement it themselves. Since the whole concept relies on components that exist on different abstraction levels, we will start from basic atomic terms that will later be enclosed in building blocks for the final solution.
We will start by explaining the notion of similarity between objects, which is essential for many machine learning problems.
Among various similarity measures, the most popular is the Euclidean distance (in fact, it is a dissimilarity measure—we will soon transform it into similarity). It is the length of a line segment between two points in Euclidean space spanned by object feature dimensions. Euclidean distance aggregates similarities between each of the features. It is defined as follows:
If two objects do not differ in any of the dimensions, the Euclidean distance is zero; otherwise, it is a positive number. Distances between all objects in a dataset are usually expressed in the form of a matrix called a distance matrix.
We will now take a closer look at one example of such a distance matrix. We will then jump from dissimilarity to similarity by transforming the distance matrix into a similarity matrix using a parameterized Gaussian similarity function. Our main goal will be to find out how to compute the similarity matrix—a structure that encodes similarities between all objects in our dataset. We will also point out all difficulties that emerge while composing it.
The Julia package named Distances.jl is worth mentioning now. Let’s install it and start looking into a distance matrix based on the iris dataset (we assume each column of a data matrix is a single observation and each row represents a single feature):
1
2
3
4
5
6
7
8
9
10
# Pkg.add("Distances")
# Pkg.add("RDatasets") # datasets source
using Distances
using RDatasets
iris = dataset("datasets", "iris")
data_matrix = transpose(convert(Array{Float64,2}, iris[:,1:4]))
# transpose: data frame is row-oriented while pairwise computes distances between columns
distance_matrix = pairwise(Euclidean(), data_matrix)
Just a quick marginal note on its speed. It is impressively fast. Try to compute a distance matrix for 10,000 objects, each having 10,000 features.
1
2
3
data_matrix_big = randn(10000,10000);
@time pairwise(Euclidean(), data_matrix_big);
# elapsed time: 13.350062394 seconds (800880240 bytes allocated, 1.11% gc time)
(Check out the benchmarks for details.)
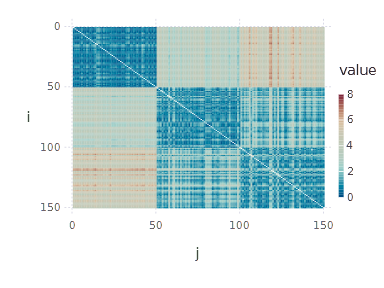
We can now visualize the iris dataset distance matrix using the Gadfly visualization package:
1
2
3
# Pkg.add("Gadfly") # install if not present
using Gadfly
spy(distance_matrix)
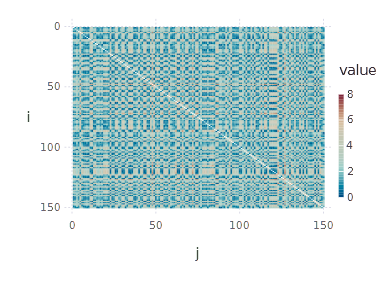
Let’s try to interpret what we see. First of all, indices
1
2
3
shuffled_data_matrix = data_matrix[:, shuffle(1:size(data_matrix,2))]
shuffled_distance_matrix = pairwise(Euclidean(), shuffled_data_matrix)
spy(shuffled_distance_matrix)
As you’ve already noticed, the Euclidean distance value reflects dissimilarity between objects, while we need a similarity measure. Therefore, we need a way to go from a distance measure to a similarity measure such that similarity = 0 reflects no similarity while similarity = 1 means practically the same objects (up to the features they are represented with).
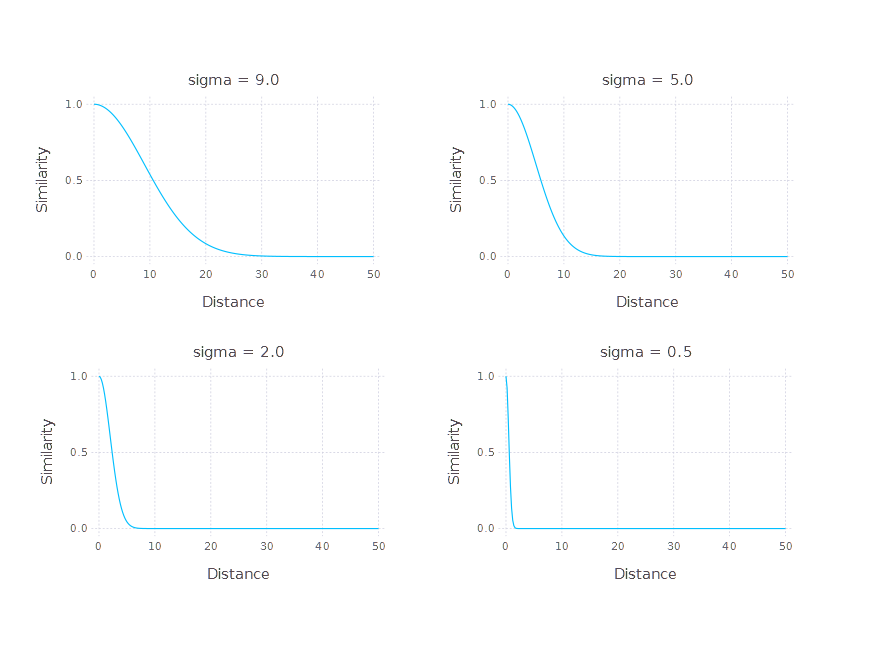
The common way of achieving this is to apply the following function called the Gaussian similarity function:
where
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
function gaussian_similarity(sigma)
return x -> exp((-x^2) / (2 * sigma^2))
end
xlab = Guide.xlabel("Distance")
ylab = Guide.ylabel("Similarity")
yvalues = Scale.y_continuous(minvalue = 0, maxvalue = 1);
plot(x=0:0.1:50, y=map(gaussian_similarity(0.5), 0:0.1:50),
Geom.line, xlab, ylab, yvalues, Guide.title("sigma = 0.5"))
plot(x=0:0.1:50, y=map(gaussian_similarity(2.0), 0:0.1:50),
Geom.line, xlab, ylab, yvalues, Guide.title("sigma = 2.0"))
plot(x=0:0.1:50, y=map(gaussian_similarity(5.0), 0:0.1:50),
Geom.line, xlab, ylab, yvalues, Guide.title("sigma = 5.0"))
plot(x=0:0.1:50, y=map(gaussian_similarity(9.0), 0:0.1:50),
Geom.line, xlab, ylab, yvalues, Guide.title("sigma = 9.0"))
The
1
2
3
4
5
color_scale = Scale.color_continuous(minvalue=0, maxvalue=1);
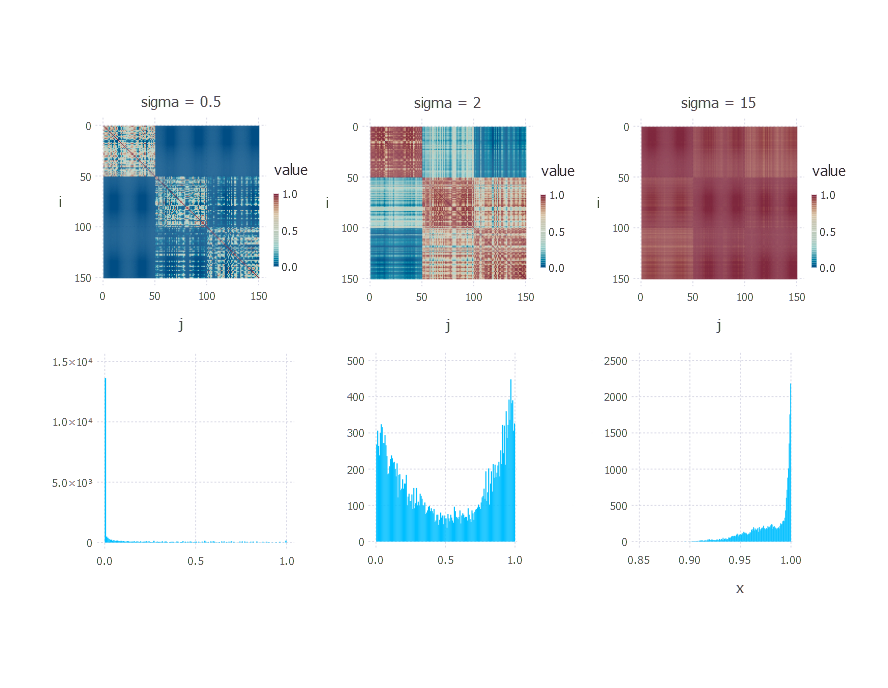
sigma = 2;
similarity_matrix = map(gaussian_similarity(sigma), distance_matrix);
spy(similarity_matrix, color_scale);
plot(x = similarity_matrix[:], Geom.histogram);
Running the code above for different values of sigma results in the following outputs:
For different choices of
Summary
We presented one approach of how to represent and encode similarities between objects in your dataset. The problem of
The next post will cover one specific interpretation of the similarity matrix. The similarity matrix will let us think of our dataset as if it were a network with a set of nodes and connections between them. This interpretation will be important for the main clustering problem (soon to be stated) we will try to solve.
Author: int8
Categories: Clustering
Tags: clustering, similarity matrix
Comments powered by Disqus.