Large Scale Spectral Clustering with Landmark-Based Representation (in Julia)
In this post we will implement and play with a clustering algorithm of a mysterious name Large Scale Spectral Clustering with Landmark-Based Representation (or shortly LSC – corresponding paper here). We will first explain the algorithm step by step and then map it to Julia code (github link).
Spectral Clustering
Spectral clustering (wikipedia entry) is a term that refers to many different clustering techniques. The core of the algorithm does not differ though. In essence, it is a method that relies on spectrum (eigendecomposition) of input data similarity matrix (or its transformations). Given input dataset encoded in a matrix
and function
One specific spectral clustering algorithm (Ng, Jordan, and Weiss 2001) relies on matrix
where
In Jordan, Weiss algorithm first
Large Scale Spectral Clustering with Landmark-Based Representation
One possible way to go to limit EVD complexity is to reduce the original problem to another (easier) one. In LSC paper authors construct matrix similar to
(authors construct
Therefore it is sufficient enough to compute Singular Value Decomposition of sparse small matrix
Construction of matrix
Matrix
where
Here is exact routine to get matrix
- Given input data matrix
- For each data point
- Normalize
Computing eigenvectors of matrix
Having
(note that it is of course different
Here authors compute matrix
Matrix
Final step: k-means on eigenvectors
Now having eigenvectors of
LSC – implementation
Let’s start with routine to get
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
using Clustering;
using Distances;
function getLandmarks(X, p, method=:Random)
if(method == :Random)
numberOfPoints = size(X,2);
landmarks = X[:,randperm(numberOfPoints)[1:p]];
return landmarks;
end
if(method == :Kmeans)
kmeansResult = kmeans(X,p)
return kmeansResult.centers;
end
throw(ArgumentError('method can only be :Kmeans or :Random'));
end
We also need a function to compute
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
function gaussianKernel(distance, bandwidth)
exp(-distance / (2*bandwidth^2));
end
function composeSparseZHatMatrix(X, landmarks, bandwidth, r)
distances = pairwise(Distances.Euclidean(), landmarks, X);
similarities = map(x -> gaussianKernel(x, bandwidth), distances);
ZHat = zeros(size(similarities));
for i in 1:(size(similarities,2))
topLandMarksIndices = selectperm(similarities[:,i], 1:r, rev=true);
topLandMarksCoefficients = similarities[topLandMarksIndices, i];
topLandMarksCoefficients = topLandMarksCoefficients / sum(topLandMarksCoefficients);
ZHat[topLandMarksIndices,i] = topLandMarksCoefficients;
end
return diagm(sum(ZHat,2)[:])^(-1/2) * ZHat;
end
Finally, we can implement core LSC routine as
1
2
3
4
5
6
7
function LSCClustering(X, nrOfClusters, nrOfLandmarks, method, nonZeroLandmarkWeights, bandwidth)
landmarks = getLandmarks(X, nrOfLandmarks, method)
ZHat = composeSparseZHatMatrix(X, landmarks, bandwidth, nonZeroLandmarkWeights)
svdResult = svd(transpose(ZHat))
clusteringResult = kmeans(transpose(svdResult[1])[:1:nrOfClusters],nrOfClusters);
return clusteringResult
end
Please note that our implementation differs a bit from the original one (here, we consider first
LSC - toy data examples
Here is the result of LSC clustering (with k-means landmark generation) on toy datasets - they all have cardinality 2000
(Datasets are available to download: cassini, shapes, smiley and spirals)
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
using Gadfly
# smiley example
smiley = readtable("smiley.csv");
smiley = transpose(convert(Array{Float64,2}, smiley[:,1:2]))
result = LSCClustering(smiley, 4, 50, :Kmeans, 4, 0.4)
plot(x = smiley[1,:], y = smiley[2,:], color = result.assignments)
# spirals example
spirals = readtable("spirals.csv");
spirals = transpose(convert(Array{Float64,2}, spirals[:,1:2]))
result = LSCClustering(spirals, 2, 150, :Kmeans, 4, 0.04)
plot(x = spirals[1,:], y = spirals[2,:], color = result.assignments)
# shapes example
shapes = readtable("shapes.csv");
shapes = transpose(convert(Array{Float64,2}, shapes[:,1:2]))
result = LSCClustering(shapes, 4, 50, :Kmeans, 4, 0.04)
plot(x = shapes[1,:], y = shapes[2,:], color = result.assignments)
# cassini example
cassini = readtable("cassini.csv");
cassini = transpose(convert(Array{Float64,2}, cassini[:,2:3]))
result = LSCClustering(cassini, 3, 50, :Kmeans, 4, 0.4)
plot(x = cassini[1,:], y = cassini[2,:], color = result.assignments)
Obviously, even though results look quite promising we cannot pass over one problem: parameter choice. Here, we have quite a few of them - number of landmarks,
LSC - MNIST dataset
Let’s finally try LSC on a ‘more serious’ dataset - handwritten digits dataset called MNIST. In this experiment we are going to compare LSC with classical k-means. We are going to use NMI (normalized mutual information) clustering quality as our metric.
Let’s quickly implement NMI (according to this)
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
# Pkg.add("Iterators")
using Iterators
function entropy(partitionSet, N)
return mapreduce(x -> -(length(x) / N) * log(length(x) / N), +, partitionSet)
end
function getPartitionSet(vector, nrOfClusters)
d = Dict(zip(1:nrOfClusters,[Int[] for _ in 1:nrOfClusters]));
for i in 1:length(vector)
push!(d[vector[i]], i)
end
return collect(values(d))
end
function mutualInformation(partitionSetA, partitionSetB, N)
anonFunc = (x) -> (
intersection = size(intersect(partitionSetA[x[1]], partitionSetB[x[2]]),1);
return intersection > 0 ?
(intersection/N) * log((intersection * N) / (size(partitionSetA[x[1]],1) * size(partitionSetB[x[2]],1))) : 0;
)
mapreduce(anonFunc, + , product(1:length(partitionSetA),1:length(partitionSetB)))
end
function normalizedMutualInformation(partitionSetA, partitionSetB, N)
mutualInformation(partitionSetA, partitionSetB, N) / ((entropy(partitionSetA, N) + entropy(partitionSetB, N)) / 2)
end
And now we are ready to go. Let’s compare our results with k-means for a sanity check (running 50 experiments):
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
# Pkg.add("MNIST")
using MNIST;
# reading dataset (60k objects)
data, labels = MNIST.traindata(); # using testdata() instead will return smaller (10k objects) dataset
# normalizing it
data = (data .- mean(data,2)) / std(data .- mean(data,2));
nrOfExperiments = 50;
kmeansResults = [];
LSCResults = [];
for i in 1:nrOfExperiments
LSCMnistResult = LSCClustering(data, 10, 350, :Kmeans, 5, 0.5);
kmeansMnistResult = kmeans(data,10);
kmeansNMI = normalizedMutualInformation(getPartitionSet(kmeansMnistResult.assignments, 10) , getPartitionSet(labels + 1 ,10), 60000)
LSCNMI = normalizedMutualInformation(getPartitionSet(LSCMnistResult.assignments, 10) , getPartitionSet(labels + 1 ,10), 60000)
push!(kmeansResults, kmeansNMI);
push!(LSCResults, LSCNMI);
end
# Pkg.add("DataFrames");
# Pkg.add("Gadfly");
using DataFrames;
using Gadfly;
df = DataFrame(nmi = [kmeansResults; LSCResults], algo = [["kmeans" for _ in 1:50];["LSC" for _ in 1:50]]);
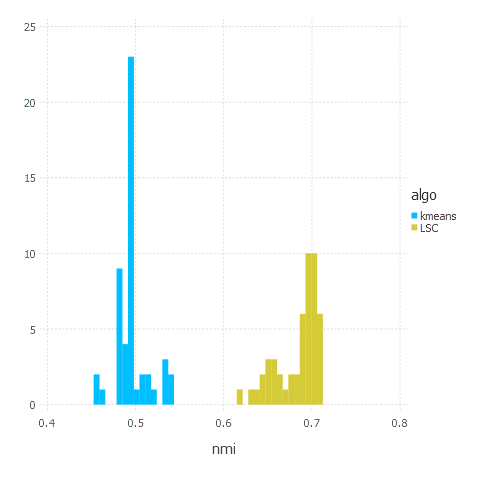
plot(df, x="nmi", color="algo", Geom.histogram(bincount=40));
Summary
That’s it. LSC clustering algorithm turned out to be better in terms of quality (as promised in a paper) than k-means. The main challenge though was proper parameters’ values choice - usually we had to tweak Gaussian kernel bandwidth and number of landmarks a bit to make the algorithm produce good results. K-means does not suffer from that problem so it is usually ‘practically’ faster - as you don’t need to experiment with parameters (which is a hard problem itself) to get good clustering.

Comments powered by Disqus.